We’re back to look at another heat map package! This is Part 2 of a 3 part series. You can find Part 1 here.
As someone who learned R using primarily the tidyverse, pheatmap had a bit of a learning curve. As a grad student, I was looking for an easier way to make a heat map with clustered data. I was trying to make heat maps for about 50 gene clusters in a programmatic way. ggplot2 wasn’t cutting it for me so I landed here. The end result was this figure & this figure, both published in this paper.
We’re going to use the same data as before. A lot of the data wrangling will look similar to Part 1.
First, we’re going to load the libraries will be using.
library(tidyverse)
library(RColorBrewer)
library(pheatmap)
Next, we’re going to read in the file using read_csv() from the tidyverse package.
df = read_csv("https://raw.githubusercontent.com/sapo83/TidyTuesday/master/2018/TT.20180403/us.avg.tuition.noCR.csv")
##
## ── Column specification ────────────────────────────────────────────────────────
## cols(
## State = col_character(),
## `2004-05` = col_character(),
## `2005-06` = col_character(),
## `2006-07` = col_character(),
## `2007-08` = col_character(),
## `2008-09` = col_character(),
## `2009-10` = col_character(),
## `2010-11` = col_character(),
## `2011-12` = col_character(),
## `2012-13` = col_character(),
## `2013-14` = col_character(),
## `2014-15` = col_character(),
## `2015-16` = col_character()
## )
df
## # A tibble: 50 x 13
## State `2004-05` `2005-06` `2006-07` `2007-08` `2008-09` `2009-10` `2010-11`
## <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 Alabama $5,683 $5,841 $5,753 $6,008 $6,475 $7,189 $8,071
## 2 Alaska $4,328 $4,633 $4,919 $5,070 $5,075 $5,455 $5,759
## 3 Arizona $5,138 $5,416 $5,481 $5,682 $6,058 $7,263 $8,840
## 4 Arkans… $5,772 $6,082 $6,232 $6,415 $6,417 $6,627 $6,901
## 5 Califo… $5,286 $5,528 $5,335 $5,672 $5,898 $7,259 $8,194
## 6 Colora… $4,704 $5,407 $5,596 $6,227 $6,284 $6,948 $7,748
## 7 Connec… $7,984 $8,249 $8,368 $8,678 $8,721 $9,371 $9,827
## 8 Delawa… $8,353 $8,611 $8,682 $8,946 $8,995 $9,987 $10,534
## 9 Florida $3,848 $3,924 $3,888 $3,879 $4,150 $4,783 $5,511
## 10 Georgia $4,298 $4,492 $4,584 $4,790 $4,831 $5,550 $6,428
## # … with 40 more rows, and 5 more variables: 2011-12 <chr>, 2012-13 <chr>,
## # 2013-14 <chr>, 2014-15 <chr>, 2015-16 <chr>
Next, we’re going to use gsub() to replace all the dollar signs & commas. lapply() allows us to apply this to every column in our data frame.
df = as.data.frame(lapply(df, function (x) {gsub("[$,]", "", x)}))
head(df)
## State X2004.05 X2005.06 X2006.07 X2007.08 X2008.09 X2009.10 X2010.11
## 1 Alabama 5683 5841 5753 6008 6475 7189 8071
## 2 Alaska 4328 4633 4919 5070 5075 5455 5759
## 3 Arizona 5138 5416 5481 5682 6058 7263 8840
## 4 Arkansas 5772 6082 6232 6415 6417 6627 6901
## 5 California 5286 5528 5335 5672 5898 7259 8194
## 6 Colorado 4704 5407 5596 6227 6284 6948 7748
## X2011.12 X2012.13 X2013.14 X2014.15 X2015.16
## 1 8452 9098 9359 9496 9751
## 2 5762 6026 6012 6149 6571
## 3 9967 10134 10296 10414 10646
## 4 7029 7287 7408 7606 7867
## 5 9436 9361 9274 9187 9270
## 6 8316 8793 9293 9299 9748
Looks good so far!
One of the quirks of pheatmap is that it will only accept a matrix with numeric values. This caused me the most trouble when I first started using this package. To handle this, we’ll apply (using sapply()) the as.numeric() function to each column except the first column. We’ll then assign this as a matrix (using as.matrix()) to a new object (df_num). Our row names for our new object are the states. We set the row names for df_num using the first column from our original data frame (df).
df_num = as.matrix(sapply(df[, 2:13], as.numeric))
rownames(df_num) = df$State
head(df)
## State X2004.05 X2005.06 X2006.07 X2007.08 X2008.09 X2009.10 X2010.11
## 1 Alabama 5683 5841 5753 6008 6475 7189 8071
## 2 Alaska 4328 4633 4919 5070 5075 5455 5759
## 3 Arizona 5138 5416 5481 5682 6058 7263 8840
## 4 Arkansas 5772 6082 6232 6415 6417 6627 6901
## 5 California 5286 5528 5335 5672 5898 7259 8194
## 6 Colorado 4704 5407 5596 6227 6284 6948 7748
## X2011.12 X2012.13 X2013.14 X2014.15 X2015.16
## 1 8452 9098 9359 9496 9751
## 2 5762 6026 6012 6149 6571
## 3 9967 10134 10296 10414 10646
## 4 7029 7287 7408 7606 7867
## 5 9436 9361 9274 9187 9270
## 6 8316 8793 9293 9299 9748
One last thing before we get to plotting our heat map: column names! The column names need to be cleaned up a bit.
We’ll used two nested gsub() commands to remove the “X” from the beginning of the string & replace the “-” with “-20”.
colnames(df_num) = gsub("[.]", "-20", gsub("X", "", colnames(df_num)))
Now, let’s plot our heat map!
pheatmap(df_num)
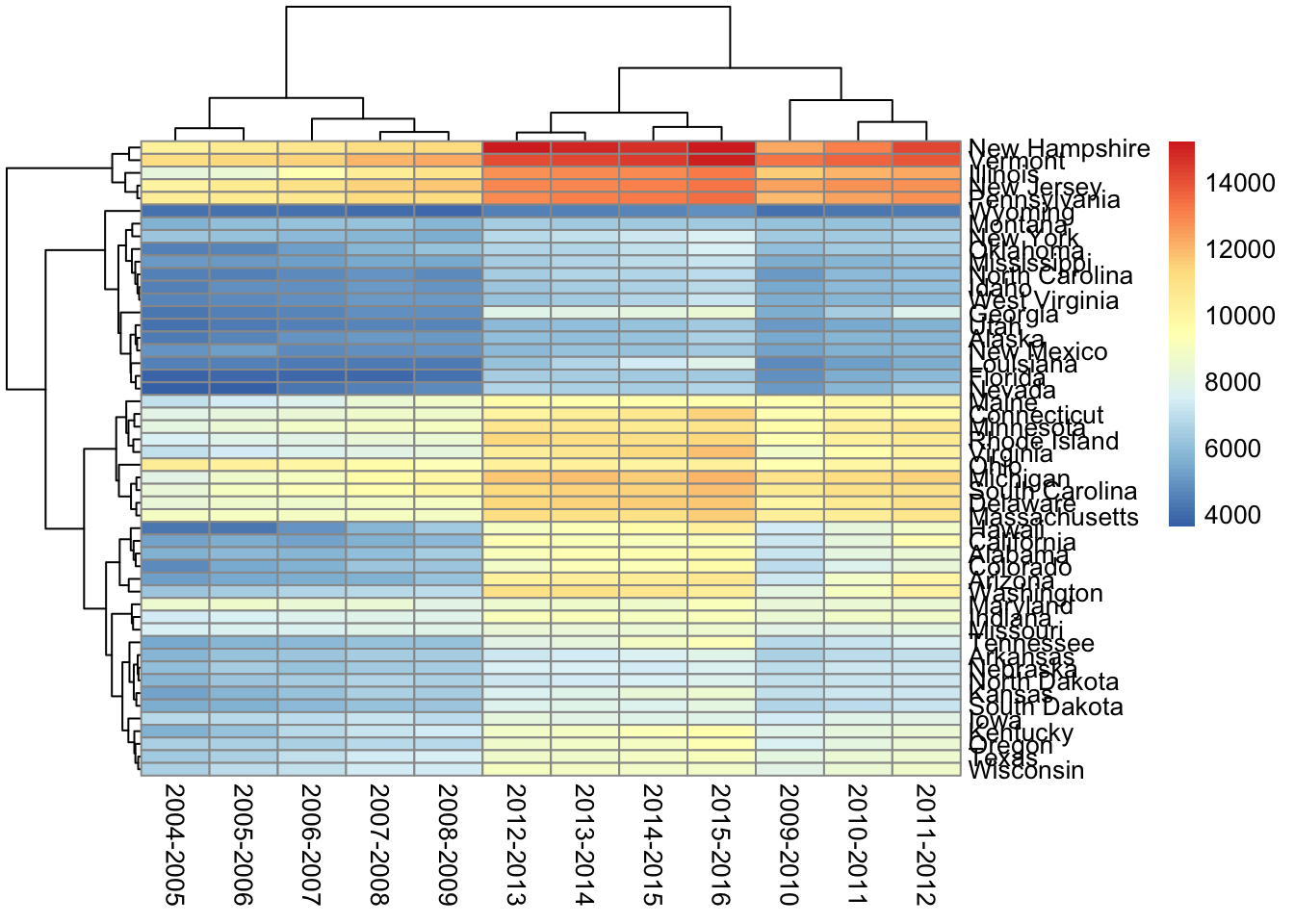
First thing I want to fix is the column order. I don’t want them clustered. I want to see them in order by date. We can add cluster_cols = FALSE to take care of that. I’ll leave the clustering on for the rows (states). With this clustering we can see which states have similar patterns in their tuition changes. This may help us see particular trends in certain regions or identify outlier(s) in a specific area.
I also want to add a color scheme to it. I’m going to stick with the color palette from Part 1.
pheatmap(df_num,
color = colorRampPalette(rev(brewer.pal(11, "RdYlBu")))(100),
cluster_cols = FALSE)
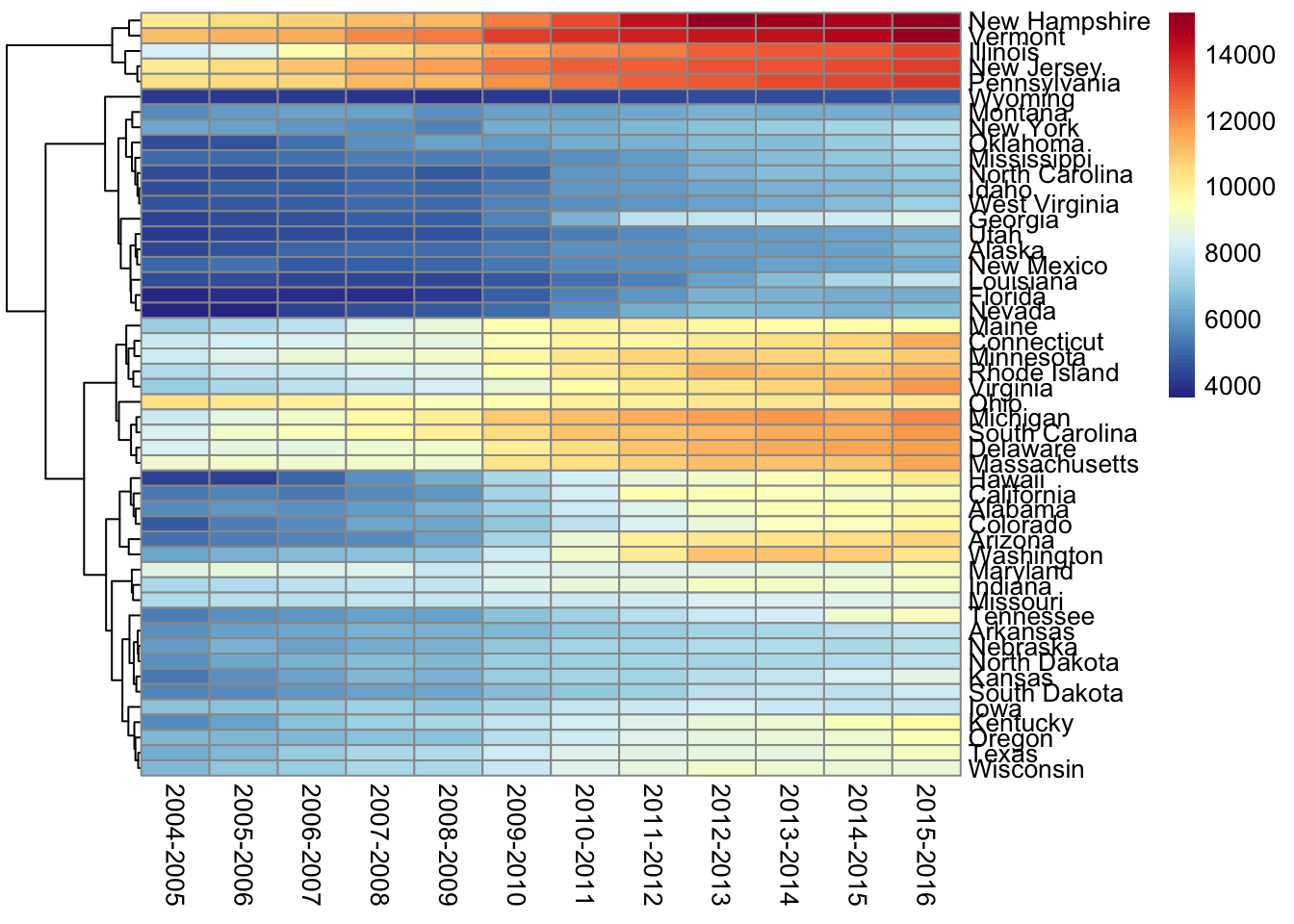
Next I want to change the cell size & remove the cell border. I’ll use NA to remove the cell border completely. I prefer squares in a heat map as opposed to rectangles so I’ll set the cell width & cell height to match.
pheatmap(df_num,
color = colorRampPalette(rev(brewer.pal(11, "RdYlBu")))(100),
cluster_cols = FALSE,
cellwidth = 10,
cellheight = 10,
border_color = NA)
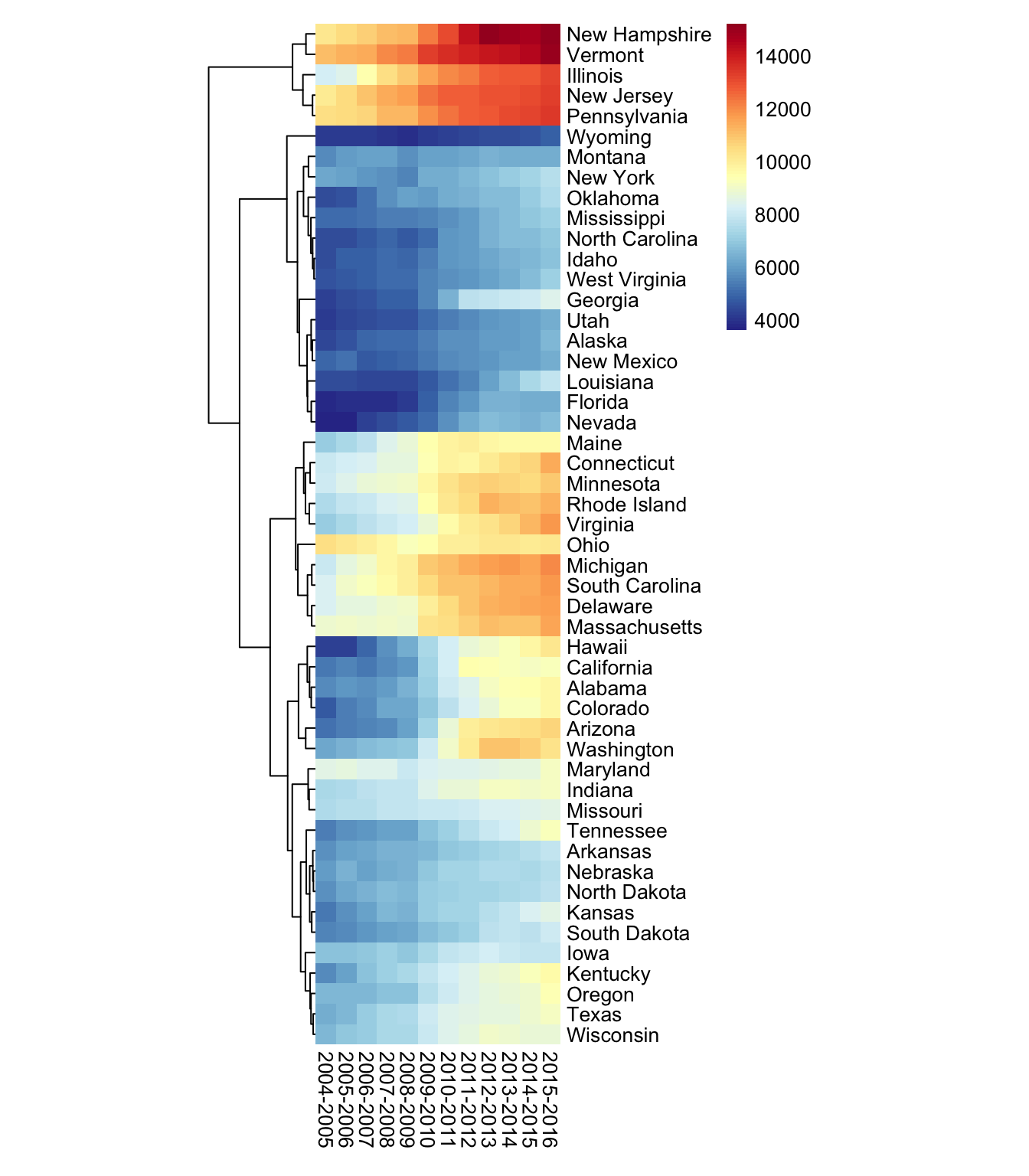
This is fairly similar to the plot in Part 1 so I’m going to stop here. Any questions/comments/concerns, I’d be happy to hear from you. You can reach out to me via Twitter or using one of the other methods on my contact page. Stay tuned for Part 3!